Data Science With Docker pt. I: Create your First Scientific Environment.

Hugo Estrada S.
Posted on March 2, 2020

Start with Data Science by creating your first scientific development environment with Docker, and learn the basis of this field using Python with Jupyter Notebooks, persisting your data in PostgreSQL and MinIO for your models, finally for dash-boarding and data visualization use Apache Superset.
First things first, the repo with all the goodies you'll need is here:
Additionally, throughout this lecture I'm going to be using Ubuntu 18.04 LTS, all my set-up configuration is tested and fully functional with this specific version of Linux.
Part 1: Why Docker <?>
Docker deploys applications and solutions in a standard unit of software that packages up code and all its dependencies so the application runs quickly and reliably from one computing environment to another. These individual solutions are called "containers".
Docker Compose allows us to deploy and run multi-container Docker applications.
We achieve this by creating and editing "YAML" or "YML" files, by defining the services, docker images, range of ports and other characteristics (such as volumes and local environment configurations).
Docker isn't a virtualization tool, it primarily focuses on automating the deployment of applications inside application containers. Application containers are designed to package and run a single service, whereas system containers are designed to run multiple processes, like virtual machines.
We're going to talk about the different images that will be part of this docker compose configuration:
• Jupyter Notebook is an interactive and easy-to-use data science environment across the most popular programming languages that doesn't only work as an IDE, but also as a presentation or education tool. For this, I picked up the scipy-notebook image that contains support for Python 3 and already has installed and preconfigured the most popular libraries for data analysis. You can find more about the official Jupyter Notebook imaged available for docker in this link:
https://jupyter-docker-stacks.readthedocs.io/en/latest/
• We need some place to store our relational and structured data, so for that purpose I've chosen PostgreSQL. It's easy to use and offers a lot of support for Python (which is very useful). The official image of PostgreSQL for docker can be found in this link: https://hub.docker.com/_/postgres
• This is a modern and easy-to use data-science environment, so we need a graphical interface for our PostgreSQL instance. I am going to use pgweg, although it does not have an official docker image, the imaged built by sosedoff it's pretty stable and is constantly maintained: https://hub.docker.com/r/sosedoff/pgweb
• Next up, we need a place to store unstructured data, like models and other type of not so common stuff. MinIO is a build high performance, cloud native data infrastructure for machine learning, analytics and application data workloads. Here's the official docker image: https://hub.docker.com/r/minio/minio/
• And last, but not least we need a dash-boarding and powerful graph tool to present, manipulate and even understand data in a visual way. The Apache Superset project offers tons of graphs, a very complex role-based control for end users and even its own SQL interface. Like pgweb, I'll use a non-official image: https://hub.docker.com/r/tylerfowler/superset
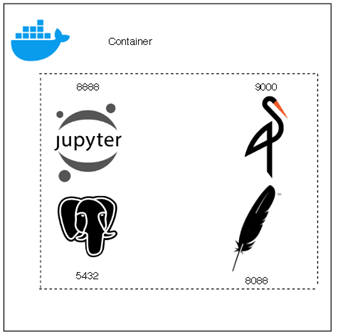
So, our multi container environment (architecturally speaking) is going to like like this:
Part 2: Connecting the Dots - Setting up our environment
It's time to start configuring all of this different components, Starting with Docker and Docker Compose.
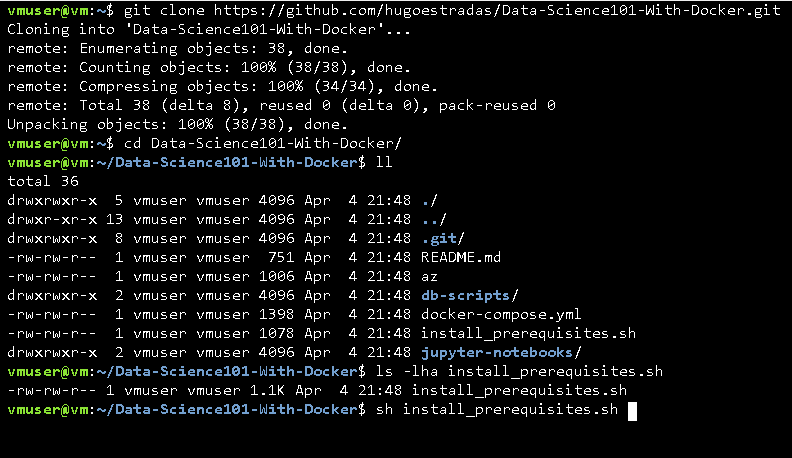
For that, I've already design an "install_prerequisites.sh" file. So if you're using Linux (as I suggest in the repository description) all you have to do is clone the repository and execute this file, with will install and configure Docker and Docker-Compose:
sh install_prerequisites.sh.
After this, you'll have Docker and Docker Compose installed in your machine.
You might as well consider granting access to these two commands for your specific user of preference (in your local environment), so you don't have to type "sudo" each time you need to run them.
sudo usermod -aG docker [your user]
Just replace "vmuser" with the name of your user, and you're ready to use the docker and docker-compose commands at will!
It's worth mention: You must reboot your local environment for the changes to take effect.
Moving on, let's try to understand how Docker works.
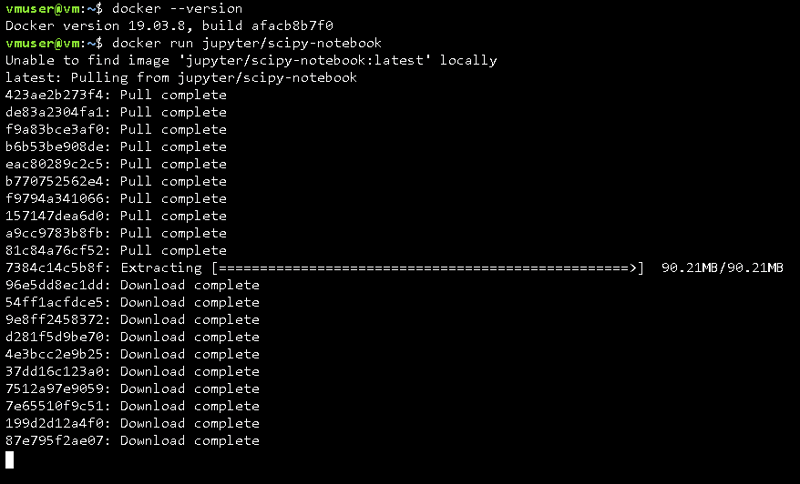
If you're interested in knowing your current version of Docker, type:
docker --version
To run your first Docker image, all you have to do is execute the "docker run" command, specifying the name of the image:
docker run [name of the image]
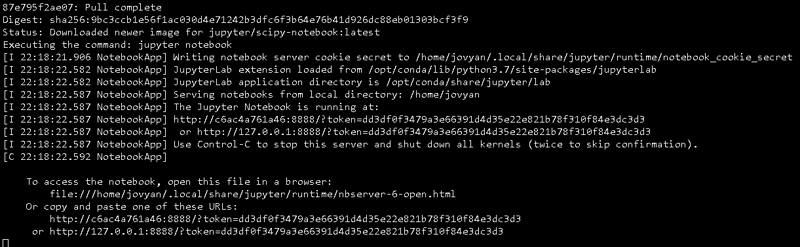
The previous command will start to download all the dependencies needed to run the specific image we indicated, this might time some time depending on the size of the image and your internet speed.
Once it's done downloading and pulling from the Docker Hub, you'll be prompted some messages in the terminal, among other things these messages indicate the URL to access the Jupyter Notebook Server.
Whoops, if you try to access to this URL you'll discover that is not possible.
That's because although the docker run command downloaded the image from the docker hub source, we never specified the port required for this purpose.
So how can we solve the problem?
The answer: mapping ports.
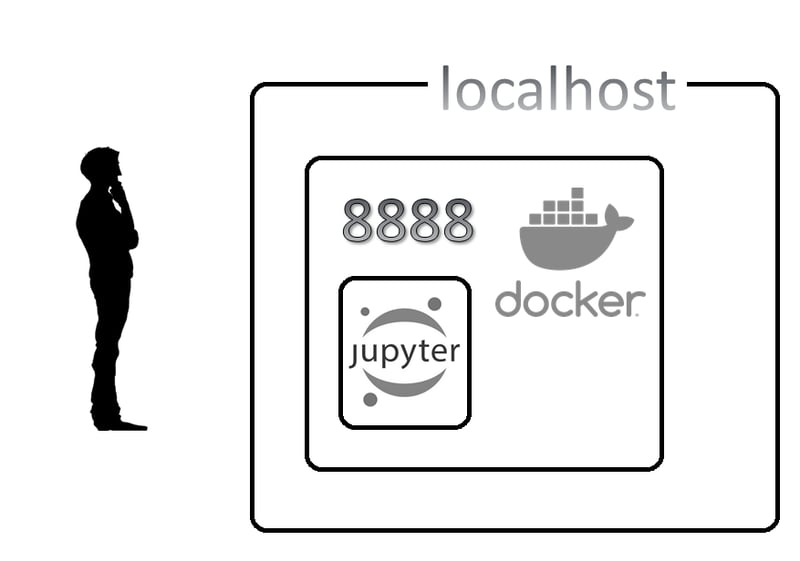
But first, let's take a look at the following diagram to understand what's
going on in the background:
In this diagram, that serious folk over there is you wondering: "why on earth I can't reach my Jupyter Image?" We also have a host machine and our docker container running our Jupyter Notebook image.
But, as I mention above, we did not explicitly indicate "hey, run this image in this port".
In other words, there is no mapping of ports.
So let's fix this. Go back to your terminal and press CTRL + c to cancel the execution of our docker image and type:
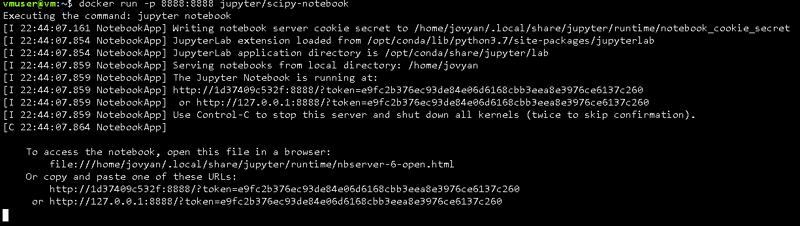
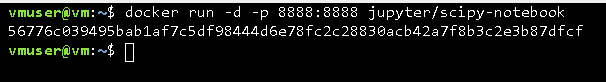
docker run -p 8888:8888 jupyter/scipy-notebook
We need to use "- p" and to indicate the port 8888.
After this, everything should look like before, but this time when you access the URL given in the terminal you'll have access to your Jupyter Server.
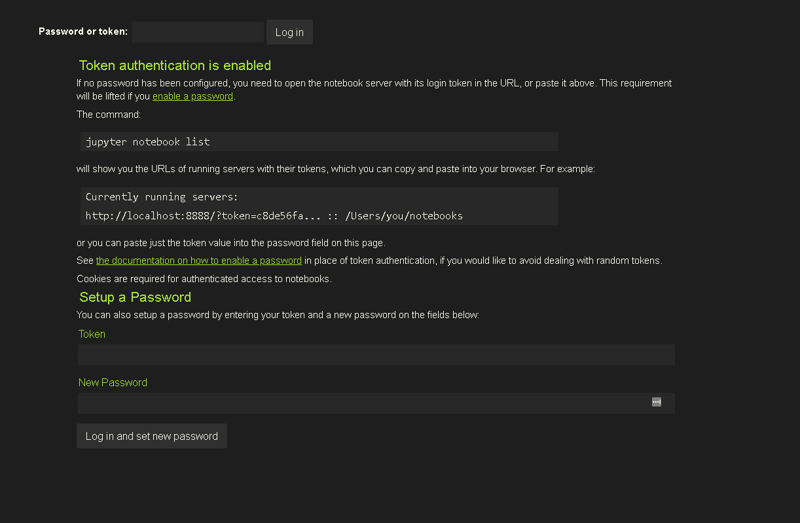
You'll be asked for an authentication token, just go back to the terminal,
select an copy the auth. token and you're ready to go!
Now, you're able to reach your Jupyter server, but you are in danger of accidentally killing your container by pressing CTRL + c or closing the terminal.
Fortunately adding the "d" flag to the command, will execute the server in the "detach mode". This will launch the container as a background task and provides the flexibility you need.
docker run -d -p 8888:8888
After doing this you will be asked again for the security token, which you are no longer able to see after running the container in detach mode.
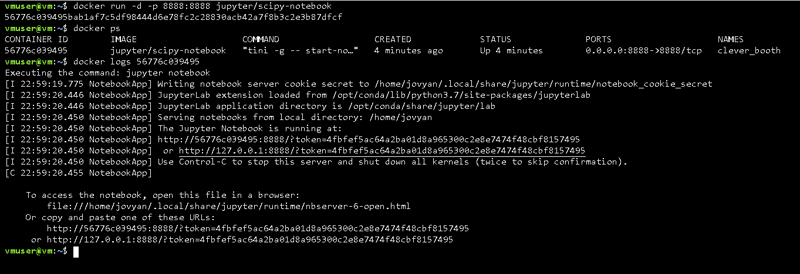
The solution for this, is to execute the "docker ps" command, which will give you the list of all the running containers, in addition to other useful information, like the container id which just so happens you need; in order to find the security token by checking the container logs with the command "docker logs"
docker ps
docker logs [container id]
So far, you've learned the basics of how to work with containers using Docker.
But the goal of this lecture, is to have a single scientific environment, using multiple docker images.
It's time to use Docker Compose. But first, let me provide a quick high-level comparison between Docker and Docker Compose.
But... what on earth is a "YAML" (or "YML") file <?>
For the Docker-Compose usage, the YAML file is a place in which we have to specify the desired infrastructure upfront. A YAML file is nothing more than a file that has hierarchical data in a human readable way.
If you want to dig more into YAML files, here you'll find useful information: https://docs.docker.com/compose/
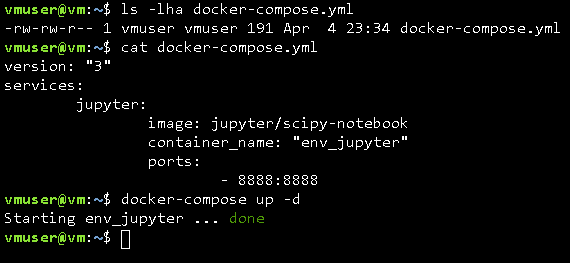
Back to our environment, here's the overall structure of a YAML file. Basically, contains two sections, the version section which specifies the docker-compose version and the services section which lists all the containers that should be included in the environment.
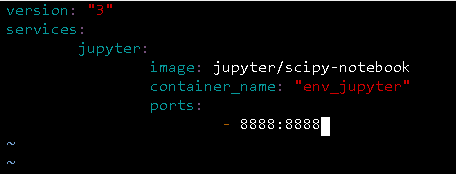
Going line by line. The first line specifies that we are using the docker-compose version 3 (which is the current version).
The second line is the beginning of the services section. At line three it's the name of the first service, in this case jupyter for Jupyter Notebook. In the line number four I'm specifying that I'd like to get the jupyter/scipy-notebook image from the docker hub. In line five I'm providing my own name for this service; hence the fact is quoted. And in lines six and seven I'm specifying the ports of this specific image.
Assuming that you store the above file as "docker-compose.yml" in your working directory, just simply launch the environment in detach mode using the following command:
docker-compose up -d
If you want to know more about the options available for the docker-compose command, here's the complete list:
https://docs.docker.com/compose/reference/
Great! you just launched your first docker-compose solution.
Now let's add another service to this file.
Part 3: Communicating the Dots - Let's make the services mutually intelligible
So far we've dug into Docker, how it works and how to deploy Docker images using the docker and docker-compose commands. Now it's time to finally define our environment. For this, we need to highly edit our yaml file. We'll add some more components to the service part, like volumes, environment and commands. If you want to know more about this, go to this link to find all the details of each section: https://docs.docker.com/compose/compose-file/
Now, let's see how to communicate between components that have been spent up with docker compose.
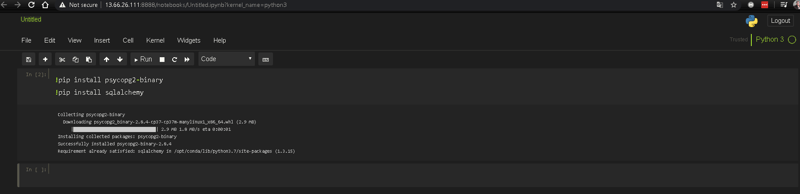
Before adding more services to our .yml file, we need to install some packages into our Jupyter Notebook, so we can load and submit data into our database.
So lets go to you Jupyter Notebook server, open up a new Python 3 file and install the following packages:
psycopg2: the most popular PostgreSQL database adapter for the Python programming language.
sqlalchemy: a Python SQL toolkit and Object Relational Mapper that gives application developers the full power and flexibility of SQL.
These two packages will allow the communication between Jupyter and PostgreSQL.
To install Python packages in Jupyter Notebooks, in the cell where you're located type the "!" symbol, which indicates that you're executing a terminal command, instead of a typical Python syntax. Then add the "pip" word, which is the default and standard package-management system used to install and manage software packages written in Python, then "install" and finally the name of the package. You can execute several installations in a single cell.
!pip install [name of the package]
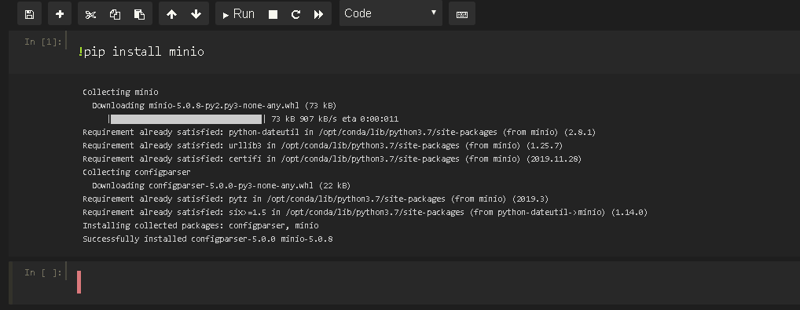
We also need to communicate Jupyter with MinIO, and for that we also need to install its own package:
Excellent, now it's time dive into the nitty-gritty details of adding the rest of our services into the environment.
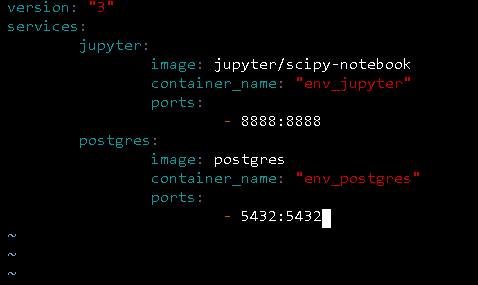
Open your yaml file docker-compose.yml and make it look like the image below:
Rhw "volume" tag, referes to directories (or files) outside of the default Union File System and exist as normal directories and files on the host filesystem. And the "environment" refers to the predefined credentials for our data persistence services MinIO and PostgreSQL.
In a nutshell, what we basically did was to add all of our services to the Docker-Compose configuration through the docker-compose.yml file.
For Jupyter Notebook, we're leaving a pretty normal configuration, the same for Apache Superset and pgweb.
The magic occurs when we define PostgreSQL, and MinIO. But why?
Well, that's because we are using an specific user and database for PostgreSQL and we're using a configured environment for MinIO.
Part 4 (optional): Put it all in an Azure VM:
If you'd like to run this environment in the Microsoft's cloud platform Azure, then follow the next steps:
First of all, you'll need to have an Azure account and an Azure subscription ready. I'm assuming you already have those two things, if not, just follow this link:
https://www.google.com/search?q=set+up+azure+account&rlz=1C1GCEU_enGT858GT858&oq=set+up+azure+account&aqs=chrome.0.0l6.2359j0j7&sourceid=chrome&ie=UTF-8
Once you have this covered, you'll need to create an Azure Cloud Shell, this procedure it's pretty straightforward, just follow the steps in this article:
https://docs.microsoft.com/en-us/azure/cloud-shell/quickstart
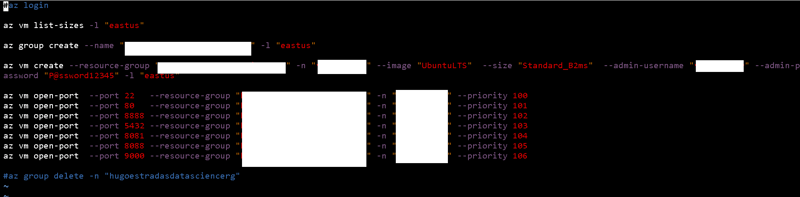
All right, in my repository locate the "az.sh" file, this is a bash file for linux. This file contains all the commands necessary to create a resource group, a virtual machine with the Ubuntu 18.04 LTS image, and open all the ports necessary for this.
Part 5: Final thoughts and conclusions.
So, if you made it to this part you successfully configured a docker-compose environment ready-to-use for data science.
You learned the basics of Docker, Docker-Compose and how to manage container solutions, how to map ports, how to install Python packages in Jupyter Notebook and how to communicate between containers in a single docker compose instance.
This containerized environment already contains all the tools and services needed to start in the Data Science field: from data analysis, to create statistical and predictive models.
This is the first lecture of a three-part series that I've created, in the next two lectures I'm going to explore the basics of Data Science using Python and the many different libraries used for data analysis, data exploration, plotting data, modeling data, some SQL basics for data science, how to create and deploy interactive dashboards used for management to provide insightful data that will make the difference in the decision making process, how create and train predictive models and many other things.
The fun does not end here!

Posted on March 2, 2020
Join Our Newsletter. No Spam, Only the good stuff.
Sign up to receive the latest update from our blog.