How do they apply BERT in the clinical domain?

Edward Ma
Posted on June 10, 2019
BERT in clinical domain
Contextual word embeddings is proven that have dramatically improved NLP model performance via ELMo (Peters et al., 2018), BERT (Devlin et al., 2018) and GPT-2 (Radford et al., 2019). Lots of researches intend to fine tune BERT model on domain specific data. BioBERT and SciBERT are introduced in last time. Would like to continue on this topic as there are another 2 research fine tune BERT model and applying in the clinical domain.
This story will discuss about Publicly Available Clinical BERT Embeddings (Alsentzer et al., 2019) and ClinicalBert: Modeling Clinical Notes and Predicting Hospital Readmission (Huang et al., 2019) while it will go through BERT detail but focusing how researchers applying it in clinical domain. In case, you want to understand more about BERT, you may visit this story.The following are will be covered:
- Building clinical specific BERT resource
- Application for ClinicalBERT
Building clinical specific BERT resource
Alsentzer et al. apply 2 millon notes in the MIMIC-III v1.4 database (Johnson et al., 2016). There are among 15 note types in total and Alsentzer et al. aggregate to either non-Discharge Summary type and Discharge Summary type. Discharge summary data is designed for downstream tasks training/ fine-tuning.

Giving that those data, ScispaCy is leveraged to tokenize article to sentence. Those sentences will be passed to BERT-Base (Original BERT base model) and BioBERT respectively for additional pre-training.
Clinical BERT is build based on BERT-base while Clinical BioBERT is based on BioBERT. Once the contextual word embeddings is trained, a signal linear layer classification model is trained for tacking named-entity recognition (NER), de-identification (de-ID) task or sentiment classification.
These models achieves a better result in MedNLI by comparing to original BERT model. Meanwhile, you may notice that there are no improvement fro i2b2 2006 and i2b2 2014 which are de-ID tasks.

Application for ClinicalBERT
In the same time, Huang et al. also focus on clinical notes. However, the major objective of Huang et al. research is building a prediction model by leveraging a good clinical text representation. Huang et al. researched that lower readmission rate is good for patients such as saving money.
Same as Alsentzer et al., MIMIC-III dataset (Johnson et al., 2016) are used for evaluation. Following same BERT practice, contextual word embeddings is trained by predicting a masked token and next sentence prediction. In short, predicting a masked token is mask a token randomly and using surrounding words to predict masked token. Next sentence prediction is a binary classifier, output of this model is classifying whether second sentence is a next sentence of first sentence or not.
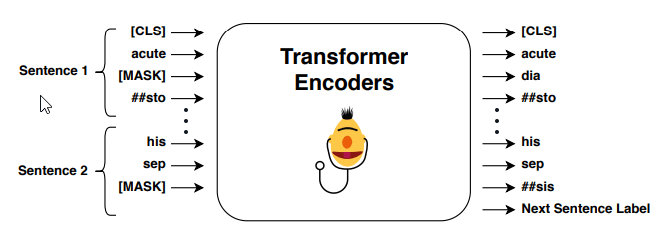
After having a pre-trained contextual word embeddings, fine-tuned process is applied on readmission prediction. It is a binary classification model to predict whether patient need to be readmission within the next 30 days.
One of the BERT model limitation is maximum length of token is 512. A long clinical note will be split to multiple parts and predicting it separately. Once all sub-part is predicted, a final probability will be aggregated. Due to the concern on using maximum or mean purely, Huang et al. combine both of them to have a accurate result.
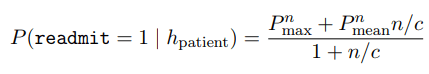
Finally, the experiment result demonstrated a fine-tuned ClinicalBERT is better than classical model.

Take Away
- Alsentzer et al. uses a signal layer of classification model to evaluate result. It may be a good start for that and expected BERT model able to learn the content. Evaluating other advanced model architecture may provide a better comprehensive experiment result.
- For long clinical note, Huang et al. uses some mathematics trick to resolve it. It may not able to capture content a very long clinical notes. May need to further think about a better way to tackle a long input.
Like to learn?
I am Data Scientist in Bay Area. Focusing on state-of-the-art in Data Science, Artificial Intelligence , especially in NLP and platform related. Feel free to connect with me on LinkedIn or following me on Medium or Github.
Extension Reading
Reference
- E. Alsentzer, J. R. Murphy, W. Boag, W. H. Weng, D. Jin, T. Naumann and M. B. A. McDermott. Publicly Available Clinical BERT Embeddings. 2019
- K. Huang, J. Altosaar and R. Ranganath. ClinicalBert: Modeling Clinical Notes and Predicting Hospital Readmission. 2019

Posted on June 10, 2019
Join Our Newsletter. No Spam, Only the good stuff.
Sign up to receive the latest update from our blog.
