Working around Glue's habit of dropping unsuspecting columns

Maurice Borgmeier
Posted on February 8, 2022

This is a story about some weird behaviour in Glue that really disappointed me. I consider it either a serious bug or terrible design decision in basic Glue functionality.
Glue ETL allows you to write serverless PySpark jobs to transform data in various formats. The Glue data catalog keeps track of which data is available in your account. It points to the actual data in different storage or database services.
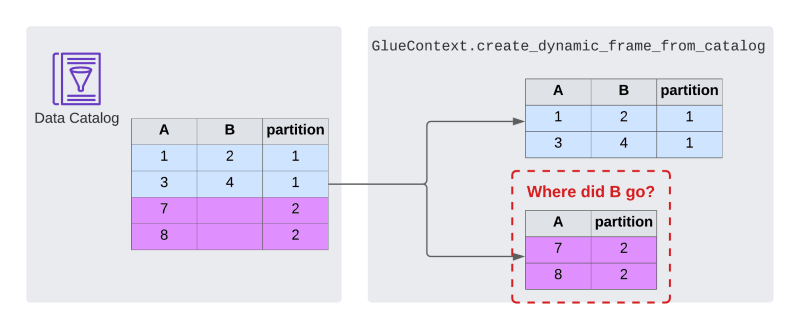
It's possible - and very common - to use the data catalog in ETL jobs to load data from data sources. It conveniently keeps track of metadata such as columns and data types. Glue provides the GlueContext as a high level abstraction to load data from the catalog. You can easily create a Glue dynamic frame from a table in the catalog using GlueContext.create_dynamic_frame_from_catalog, which you can later turn into a common PySpark data frame.
I've used this functionality many times and didn't notice any significant problems for the longest time. That changed in a recent project. I was working with a partitioned table and tried to load it into a data frame. That worked well in one partition, but led to problems in another. When I read the second partition, the data frame was missing columns that the first one from the same table had.
I was confused. That was not supposed to happen. After the initial confusion was over, I was looking for the culprit. Initially I suspected the crawler, because it had annoyed me many times before. Checking the table metadata in the data catalog revealed nothing suspicious. The base table had all the columns and so did the partition definitions. In fact the configurations were pretty much identical. I also checked the underlying data. It turned out that the data for partition #2 contained the missing column B - but the column was empty for all rows.
That shouldn't make a difference. Empty columns are not uncommon and shouldn't be discarded, because processes can rely on them, e.g. to fill them with data. This left me with the hypothesis that something was discarding empty columns without telling anyone.
Now the question was if it's Glue's or PySpark's fault. By listing the columns in the dynamic frame, I realized that the columns were missing already. That makes it clear that Glue is to blame. It also turns out that I'm not the only one to notice, there's a stackoverflow-question talking about this problem.
Glue silently drops empty columns when it reads a table or partition from the data catalog.
I can't for the life of me figure out why somebody would think this an okay behaviour. It's not configurable by the way. Glue just decides which columns of your table you'll get to use.
Arguably empty columns aren't the worst thing to lose, it would be worse if it dropped columns that hold data. That's beside the point though. Why should Glue decide to silently drop columns I might want to use without giving me an option to change that behaviour?
In the process I was building at the time, I still needed the columns even though they may be empty for a couple of partitions. That's why I decided to write a wrapper function that adds any missing columns from the base table to the data frame we get from Glue. It also sets the correct column data types even though the column itself only contains None/null values.
I achieve this by describing the table using PySpark to get the meta information about the table. This shows even columns that Glue later drops. We can then use this data to add empty columns with the correct data type to the data frame.
import pyspark.sql.functions as f
from awsglue.context import GlueContext
from pyspark.context import SparkContext
from pyspark.sql import SQLContext
def read_table_from_catalog(
database: str,
table_name: str,
push_down_predicate: str = None
):
"""
Returns a data frame from the Glue Data Catalog and makes sure all the columns
"""
spark_context = SparkContext.getOrCreate()
sql_context = SQLContext(spark_context)
df_table_info = sql_context.sql(f"describe table {database}.{table_name}")
# Get all the columns in the table with their data type
# We have to filter a little bit, the output is messy
df_table_info_filtered = df_table_info.filter(
f.col("col_name").startswith("#") == False
).distinct().select(
f.col("col_name").alias("name"),
f.col("data_type").alias("type"),
)
# Convert this to a dictionary
column_data_type_list = map(lambda row: row.asDict(), df_table_info_filtered.collect())
column_to_datatype = { item["name"]: item["type"] for item in column_data_type_list}
kwargs = {
"database": database,
"table_name": table_name,
}
if push_down_predicate is not None:
kwargs["push_down_predicate"] = push_down_predicate
# Get a dynamic frame from Glue
glue_context = GlueContext(spark_context)
dy_frame = glue_context.create_dynamic_frame_from_catalog(
**kwargs
)
df = dy_frame.toDF()
# Get a dictionary of the missing columns we need to add
columns_to_add = {
key: value for key, value in column_to_datatype.items()
if key not in df.columns
}
# Construct empty columns with the correct data type
additional_empty_columns = [
f.lit(None).cast(col_datatype).alias(col_name)
for col_name, col_datatype in columns_to_add.items()
]
df = df.select(
df["*"],
*additional_empty_columns,
)
return df
Here's a link to the repo I used to build this that contains a way to reproduce the problem. I also talked to AWS support about this (CASE 9522467631) and the very friendly support representative was able to reproduce the problem with my sample data and created a ticket for the service team. A workaround they suggested was to read the data directly from S3, but then you lose all the benefits of the data catalog with the data types, partitions etc. so that option has many drawbacks.
So here is my workaround for a problem that really shouldn't exist. Making this configurable is this weeks' wish for the #awswishlist. I'd appreciate it if they also change the default to reading all columns, but since that would break an undocumented API behaviour I doubt this is going to happen.
— Maurice

Posted on February 8, 2022
Join Our Newsletter. No Spam, Only the good stuff.
Sign up to receive the latest update from our blog.